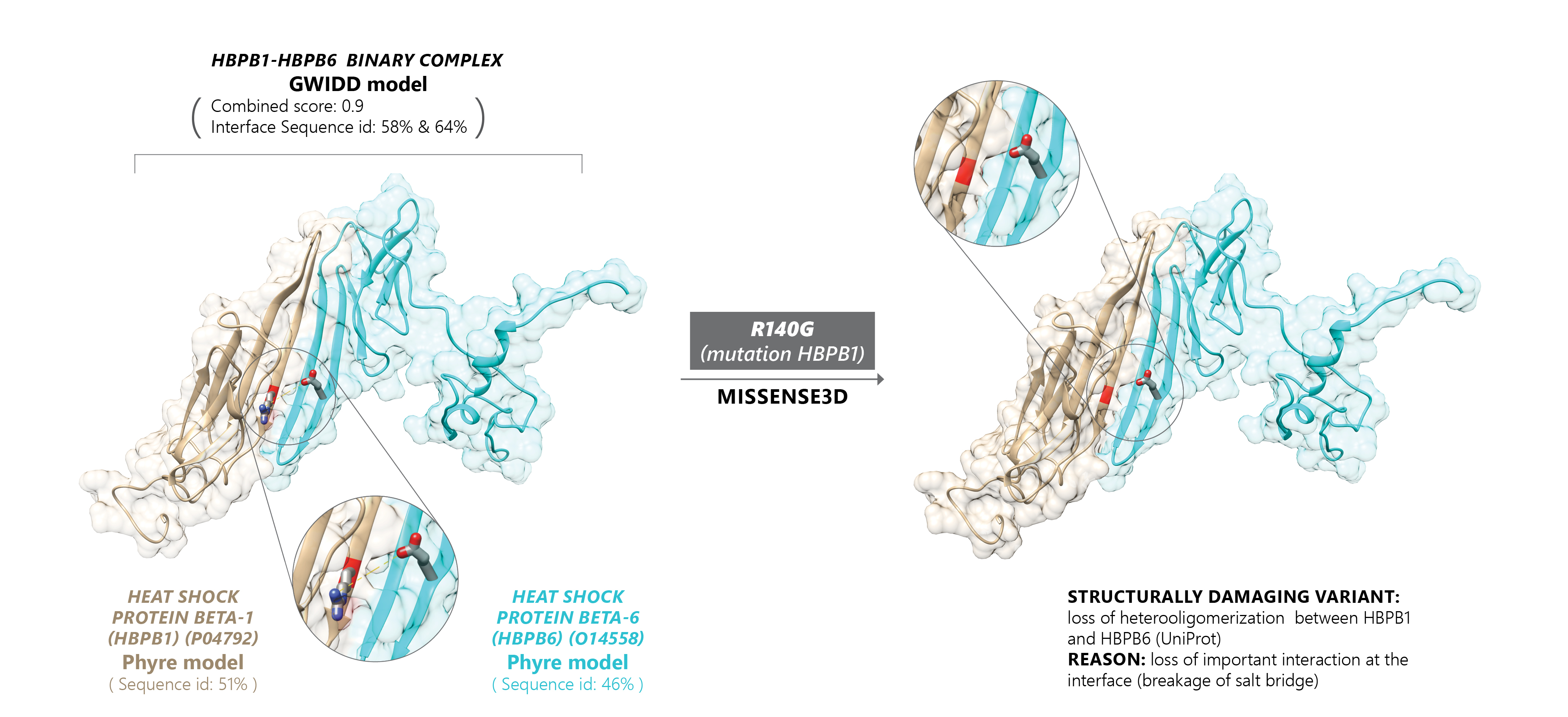
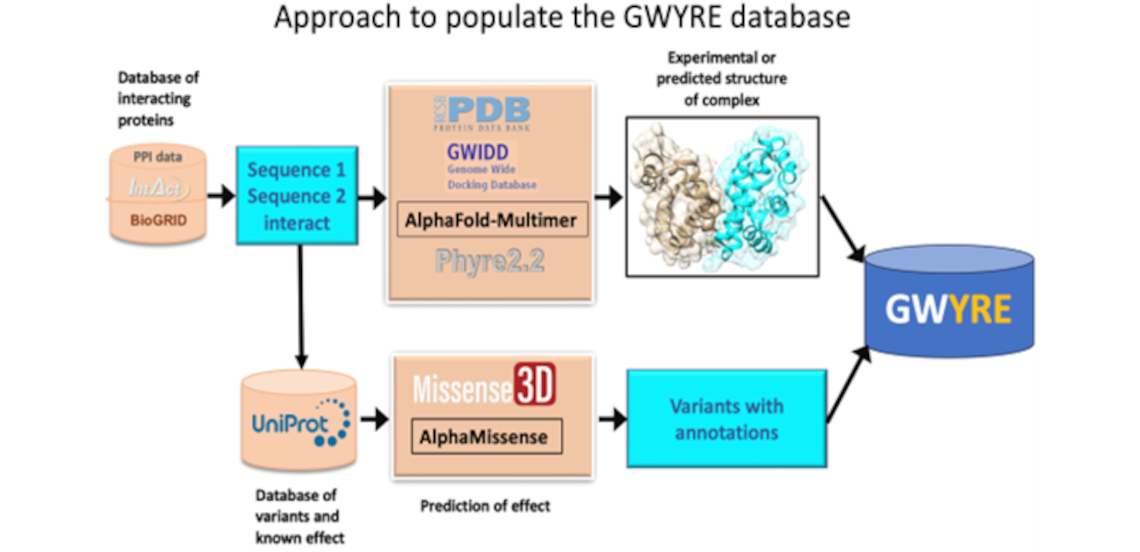
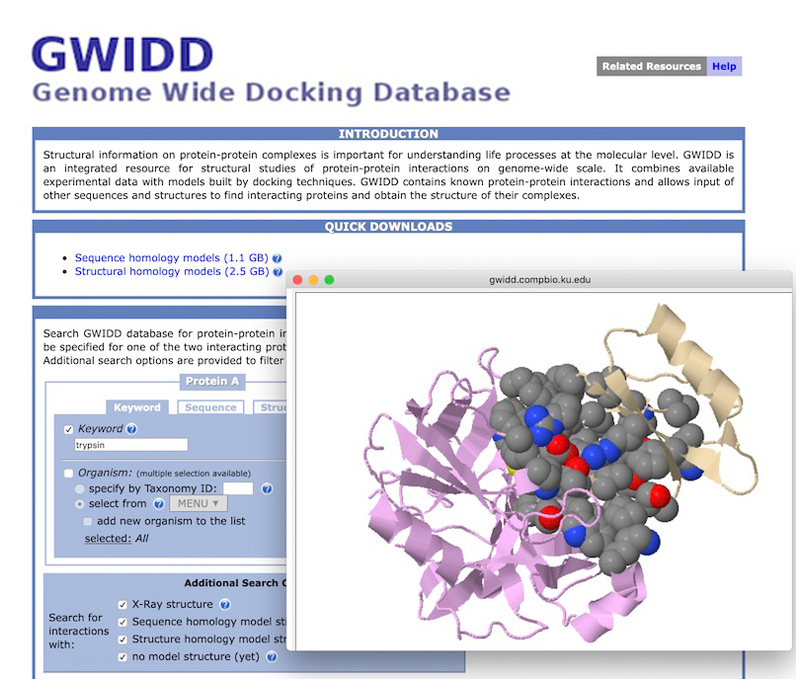
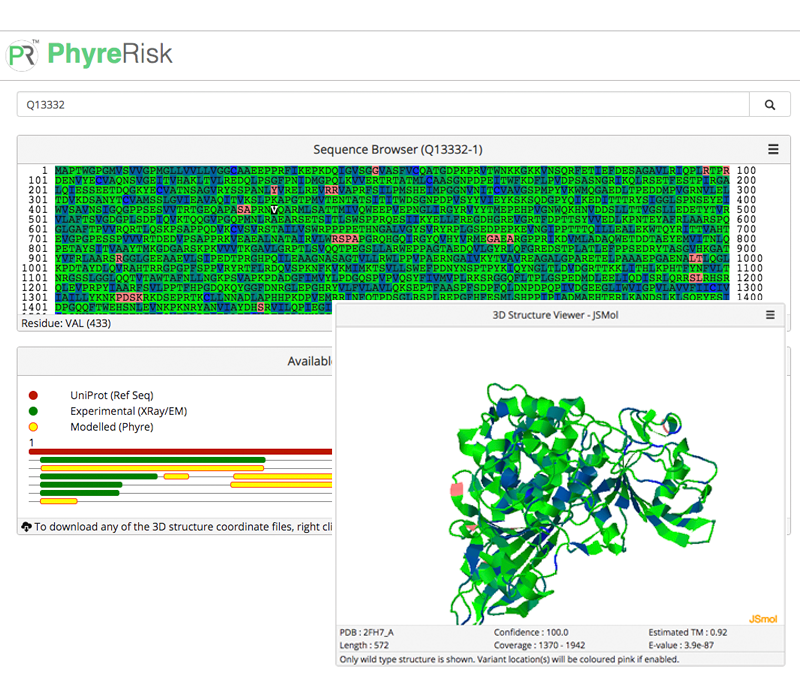
The project offers a comprehensive resource of quaternary structures (experimental and/or modeled by AI-based AlphaFold) of experimentally verified protein-protein interactions (derived from the IntAct and BioGRID databases). It also includes an assessment of the phenotypic effects of genetic mutations with a special focus on amino acids substitutions on and in the nearest vicinity of protein-protein interfaces. Both chains of all binary complexes, currently processed in GWYRE, are either extracted from the corresponding PDB file (denoted as experimental on the Search and Visualization page) or modeled as one entity by AlphaFold version 2.3 (Alphafold2 fold-and-dock model).
Search and Download
Search tool offers finding all binary complexes associated with specific protein/gene by entering whole/part of protein/gene ID(s) or name(s). Search results can be visualized by each pair of the chains in the search results with the opportunity to explore multiple interfaces for the same protein pair as well as variant information mapped on the specific interface (if available). Download tool offers a structure file in the cif (for experimental structures) of PDB format (model structures) along with a text file with the relevant information.
Downloads
GWYRE 2 complexes
[File Size: 8.8GB]
GWYRE 1 complexes
[File Size: 5.3GB]
Database Status
| Type of GWYRE Entry | Source of GWYRE Entry | Number of GWYRE entries |
|---|---|---|
| Experimental | PDB | 9048 |
| Model | Alphafold2-Multimer | 37921 |
Related Resources
Contact us
Center for Computational Biology
University of Kansas, Lawrence, KS 66045, USA.
vakser@ku.edu
Centre for Integrative Systems Biology and Bioinformatics (CISBIO)
Imperial College London, London. SW7 2AZ, UK.
m.sternberg@imperial.ac.uk